Topic 5 | Genomes and Regulation
📅 Date: 06/07 October 2025
📖 Synopsis: Exploring gene structure and annotations using the Ensembl genome browser
Lecture topics
- Navigating the Ensembl genome browser — Ensembl
- Exploring the Glucagon-like peptide 1 receptor (GLP1R) gene — Ensembl
- Understanding gene structure (exons, introns, UTRs, CDS) — Ensembl
Learning goals
- Learn to locate a gene in a genome browser.
- Retrieve basic genomic information such as chromosomal position and gene structure.
- Interpret visual representations of genes and transcripts.
TP Activity 5:
Exploring the Semaglutide Target - GLP1R in Ensembl
Goal
Explore and critically interpret bioinformatics resources to connect semaglutide’s drug target GLP1R gene with its gene features and annotations.
Use the following database:
How to work
Search for semaglutide in DrugBank and skim the drug overview.
In the drug page, go to the Targets section and open the UniProt entry for the human target (GLP1R).
In UniProt, use the External links tab and find Genome annotation databases. Click the Ensembl gene link (the ID that begins with ENSG).
About Ensembl IDs
Ensembl identifiers start with ENS:
• ENSG = gene (e.g., the GLP1R gene)
• ENST = transcript
• ENSP = protein
• ENSE = exon
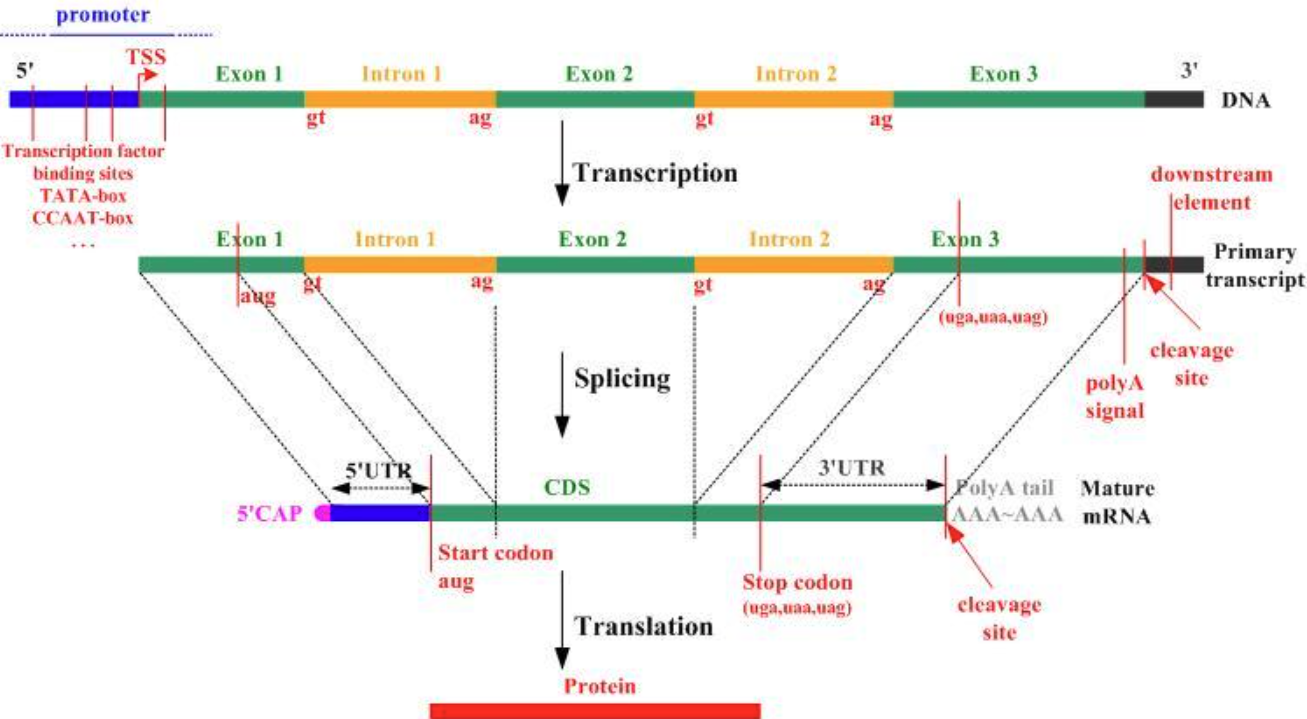
• …In Ensembl (gene entry for GLP1R), answer:
Where is the GLP1R gene located?
- Chromosome, genomic coordinates, genome assembly version, and strand.
How many transcripts are annotated for GLP1R, and what are their Ensembl transcript IDs (ENST)?
- Identify the canonical and/or MANE Select transcript if shown.
For the longest protein-coding transcript, how many exons and introns does it have?
What are the coordinates of all exons for that transcript?
What are the coordinates of the 5′ UTR and 3′ UTR?
What are the coordinates of the Transcription Start Site (TSS) and the start codon?
Reminder: the start codon encodes Methionine (ATG). The TSS is the first transcribed base of the transcript and may be upstream of the start codon.
What is the total gene length in base pairs (bp)?
How would you compute the total Coding Sequence (CDS) length?
Remember:
feature length = end − start + 1.
Tip: Ensembl coordinates are 1-based, inclusive; always confirm the strand.